Enhanced comment feature has been enabled for all readers including those not logged in. Click on the Discussion tab (top left) to add or reply to discussions.
Genetic Evaluation: Difference between revisions
| Line 19: | Line 19: | ||
</math> | </math> | ||
</center> | </center> | ||
When the subject is genetic evaluation, one of the most common questions is “What is the main difference among ssGBLUP, BLUP, and genomic BLUP (GBLUP)?” In a nutshell, ssGBLUP uses phenotypes, pedigree, and genotypes for both genotyped and non-genotyped animals, whereas BLUP uses phenotypes and pedigree for all animals and GBLUP uses phenotypes and genotypes only for genotyped animals | When the subject is genetic evaluation, one of the most common questions is “What is the main difference among ssGBLUP, BLUP, and genomic BLUP (GBLUP)?” In a nutshell, ssGBLUP uses phenotypes, pedigree, and genotypes for both genotyped and non-genotyped animals, whereas BLUP uses phenotypes and pedigree for all animals and GBLUP uses phenotypes and genotypes only for genotyped animals. | ||
In the US, ssGBLUP has been used for genomic evaluation of beef and dairy cattle, pigs, chickens, and fish. For more information about ssGBLUP for beef cattle evaluation check Lourenco et al. (2015)<ref> Lourenco, D. A. L., S. Tsuruta, B. O. Fragomeni, Y. Masuda, I. Aguilar, A. Legarra, J. K. Bertrand, T. Amen, L. Wang, D. W. Moser, and I. Misztal. 2015. Genetic evaluation using single-step genomic best linear unbiased predictor in American Angus. Journal of Animal Science 93: 2653-2662.</ref> and Misztal & Lourenco (2018) <ref> Misztal, I. and D. Lourenco. 2018. Current research in unweighted and weighted ssGBLUP. In Proc. Beef Improvement Federation 11th genetic prediction workshop 11:6-13. </ref>. | In the US, ssGBLUP has been used for genomic evaluation of beef and dairy cattle, pigs, chickens, and fish. For more information about ssGBLUP for beef cattle evaluation check Lourenco et al. (2015)<ref> Lourenco, D. A. L., S. Tsuruta, B. O. Fragomeni, Y. Masuda, I. Aguilar, A. Legarra, J. K. Bertrand, T. Amen, L. Wang, D. W. Moser, and I. Misztal. 2015. Genetic evaluation using single-step genomic best linear unbiased predictor in American Angus. Journal of Animal Science 93: 2653-2662.</ref> and Misztal & Lourenco (2018) <ref> Misztal, I. and D. Lourenco. 2018. Current research in unweighted and weighted ssGBLUP. In Proc. Beef Improvement Federation 11th genetic prediction workshop 11:6-13. </ref>. | ||
Revision as of 18:55, 21 February 2019
EPD
Utility (compared to actual/adjusted phenotypes, ratios, disjoined marker scores, etc.) (Suggested writer: Megan Rolf)
Basic Models
BLUP
ssGBLUP (Suggested writer: Daniela Lourenco)
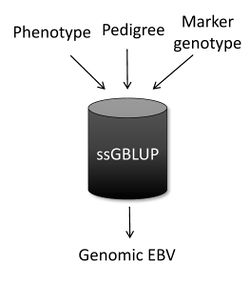
Single-step genomic BLUP (ssGBLUP) [1][2] is a method developed to enable the inclusion of marker genotypes into the well-known BLUP machinery. The idea of ssGBLUP came from the fact that only a small portion of the animals in the pedigree is genotyped. In this way, one approach to account for all animals (i.e., genotyped and non-genotyped) in the evaluation would be to combine pedigree and genomic relationships and use this as the covariance structure in the BLUP mixed model equations. Thus, ssGBLUP uses marker information to construct genomic relationships.
Legarra et al. (2009)[3] stated that genomic evaluations would be simpler if genomic relationships were available for all animals in the model. Then, their idea was to look at the pedigree relationship as a priori relationship and at the genomic relationship as the observed relationship. Based on that, they showed the genomic information could be extended (i.e., imputed) to non-genotyped animals. This means that in ssGBLUP pedigree relationships for non-genotyped animals are enhanced by the genomic information of their relatives. As a result, the output of ssGBLUP for each animal is automatically a genomic EBV.
The genomic EPD is then calculated as:
When the subject is genetic evaluation, one of the most common questions is “What is the main difference among ssGBLUP, BLUP, and genomic BLUP (GBLUP)?” In a nutshell, ssGBLUP uses phenotypes, pedigree, and genotypes for both genotyped and non-genotyped animals, whereas BLUP uses phenotypes and pedigree for all animals and GBLUP uses phenotypes and genotypes only for genotyped animals.
In the US, ssGBLUP has been used for genomic evaluation of beef and dairy cattle, pigs, chickens, and fish. For more information about ssGBLUP for beef cattle evaluation check Lourenco et al. (2015)[4] and Misztal & Lourenco (2018) [5].
Marker effects in ssGBLUP
Although the marker information in ssGBLUP is used to construct genomic relationships, it is possible to calculate SNP effects once we obtain genomic EBV (Wang et al., 2012[6], Lourenco et al., 2015[7]). Marker effects can be then used to calculate predictions based only on marker genotypes for young genotyped animals that are not yet or will never make into an official evaluation.
Single-step Hybrid Marker Effects Models (Suggested writer: Bruce Golden)
Interim Calculations
Bias
(in)complete reporting / contemporary groups / preferential treatment (Suggested writer: Bob Weaber
Accuracy (Suggested writer: Matt Spangler)
meaning of accuracy
what impacts accuracy
different definitions of accuracy (true, BIF, reliability)
Variance components (Suggested writer: Steve Kachman)
Impact on EPD, accuracy, genetic gain (Suggested writer: Steve Kachman)
Heterogeneous variance
Connectivity (Suggested writer: Ron Lewis)
Measures of (Suggested writer: Ron Lewis)
==Impact on GE== (Suggested Writer: Ron Lewis)
Current GE
How each breed (organization) is modeling each trait (Suggested writers: Steve Miller, Lauren Hyde, AHA)
- ↑ Legarra, A., I. Aguilar, and I. Misztal. 2009. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 92:4656-4663.
- ↑ Aguilar, I., I. Misztal, D. L. Johnson, A. Legarra, S. Tsuruta, and T. J. Lawlor. 2010. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. Journal of Dairy Science 93: 743-752.
- ↑ Legarra, A., I. Aguilar, and I. Misztal. 2009. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 92:4656-4663.
- ↑ Lourenco, D. A. L., S. Tsuruta, B. O. Fragomeni, Y. Masuda, I. Aguilar, A. Legarra, J. K. Bertrand, T. Amen, L. Wang, D. W. Moser, and I. Misztal. 2015. Genetic evaluation using single-step genomic best linear unbiased predictor in American Angus. Journal of Animal Science 93: 2653-2662.
- ↑ Misztal, I. and D. Lourenco. 2018. Current research in unweighted and weighted ssGBLUP. In Proc. Beef Improvement Federation 11th genetic prediction workshop 11:6-13.
- ↑ Wang, H., I. Misztal, I. Aguilar, A. Legarra, and W. M. Muir. 2012. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 94(2):73-83.
- ↑ Lourenco, D. A. L., S. Tsuruta, B. O. Fragomeni, Y. Masuda, I. Aguilar, A. Legarra, J. K. Bertrand, T. Amen, L. Wang, D. W. Moser, and I. Misztal. 2015. Genetic evaluation using single-step genomic best linear unbiased predictor in American Angus. Journal of Animal Science 93: 2653-2662.