Enhanced comment feature has been enabled for all readers including those not logged in. Click on the Discussion tab (top left) to add or reply to discussions.
Gene Edited Animal Data: Difference between revisions
No edit summary |
|||
| Line 47: | Line 47: | ||
* ''Description of the intended alteration (e.g. gene targeted, location in the genome, and intended alteration e.g. allele substitution or SNP change) | * ''Description of the intended alteration (e.g. gene targeted, location in the genome, and intended alteration e.g. allele substitution or SNP change) | ||
* ''Details of how the intended alteration was achieved (e.g. editing of somatic cells followed by cloning, or introduction of editing regents into zygote and which editing method) | * ''Details of how the intended alteration was achieved (e.g. editing of somatic cells followed by cloning, or introduction of editing regents into zygote and which editing method) | ||
* ''Timeline of when the | * ''Timeline of when the intended alteration(s) were made and by which entity (e.g. company or laboratory) | ||
* ''Data confirming the intended alteration occurred (e.g. Sanger sequencing of targeted genomic region showing knockout or knock-in) showing that the | * ''Data confirming the intended alteration occurred (e.g. Sanger sequencing of targeted genomic region showing knockout or knock-in) showing that the intended alteration(s) that were actually made | ||
* ''If a donor template plasmid was used in the editing process data should be obtained to confirm the presence or absence of the editing regents in the genome (i.e. data as to whether there was integration of the plasmid backbone into the genome) | * ''If a donor template plasmid was used in the editing process data should be obtained to confirm the presence or absence of the editing regents in the genome (i.e. data as to whether there was integration of the plasmid backbone into the genome) | ||
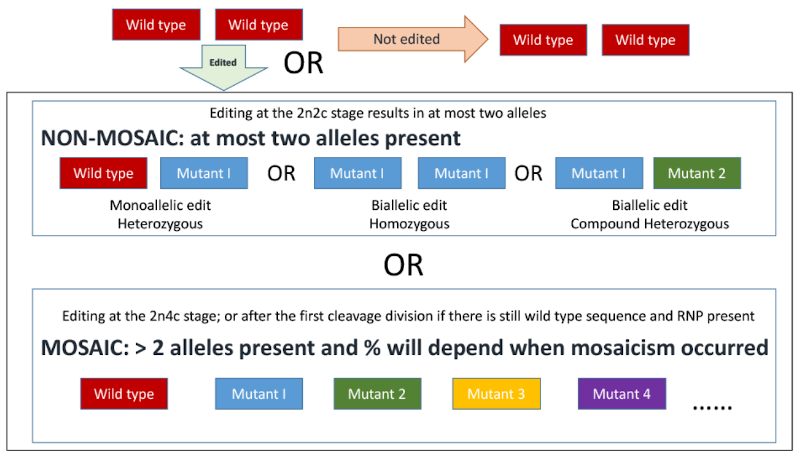
* ''Data documenting whether the animal is heterozygous monollelic edit, a homozygous biallelic edit, a compound heterozygous biallelic edit, or mosaic for the intended alteration (See Figure 2) | * ''Data documenting whether the animal is heterozygous monollelic edit, a homozygous biallelic edit, a compound heterozygous biallelic edit, or mosaic for the intended alteration (See Figure 2) | ||
* ''The entity performing the editing should store the information they used to make the animal, and confirm the | * ''The entity performing the editing should store the information they used to make the animal, and confirm the intended alteration(s) and should provide a certificate to the breeder/breed association confirming the genotype of the animal (e.g. homozygous biallelic polled (PP), heterozygous monoallelic slick (Ss)). | ||
* ''If an animal is mosaic it would be useful to know if it is germline mosaic as this can affect expected Mendelian inheritance ratios in the offspring. This can take some time as it requires the animal to sexually mature and either test semen or produce offspring, and so perhaps what is important is that breeders know the animal is mosaic so they are aware of this possibility.'' | * ''If an animal is mosaic it would be useful to know if it is germline mosaic as this can affect expected Mendelian inheritance ratios in the offspring. This can take some time as it requires the animal to sexually mature and either test semen or produce offspring, and so perhaps what is important is that breeders know the animal is mosaic so they are aware of this possibility.'' | ||
<center> | <center> | ||
| Line 57: | Line 57: | ||
</center> | </center> | ||
'''Figure 2.''' Schematic representation of possible outcomes from CRISPR-mediated mutation by cytoplasmic injection of an in vitro fertilized zygote (one cell embryo). 2n = number of homologous chromosomes, i.e. diploid. 2c/4c = number of copies of chromosomes either before or after DNA replication. Image from Supplementary materials of Hennig et al., 2020<ref>Hennig, S.L., Owen, J.R., Lin, J.C., Young, A.E., Ross, P.J., Van Eenennaam, A.L., Murray, J.D., 2020. Evaluation of mutation rates, mosaicism and off target mutations when injecting Cas9 mRNA or protein for genome editing of bovine embryos. Scientific Reports 10, 22309. doi:10.1038/s41598-020-78264-8.</ref>. | '''Figure 2.''' Schematic representation of possible outcomes from CRISPR-mediated mutation by cytoplasmic injection of an in vitro fertilized zygote (one cell embryo). 2n = number of homologous chromosomes, i.e. diploid. 2c/4c = number of copies of chromosomes either before or after DNA replication. Image from Supplementary materials of Hennig et al., 2020<ref>Hennig, S.L., Owen, J.R., Lin, J.C., Young, A.E., Ross, P.J., Van Eenennaam, A.L., Murray, J.D., 2020. Evaluation of mutation rates, mosaicism and off target mutations when injecting Cas9 mRNA or protein for genome editing of bovine embryos. Scientific Reports 10, 22309. doi:10.1038/s41598-020-78264-8.</ref>. | ||
=References= | =References= | ||
Revision as of 23:28, 18 September 2021
THIS IS A DRAFT ARTICLE PENDING APPROVAL
Genome or Gene Editing
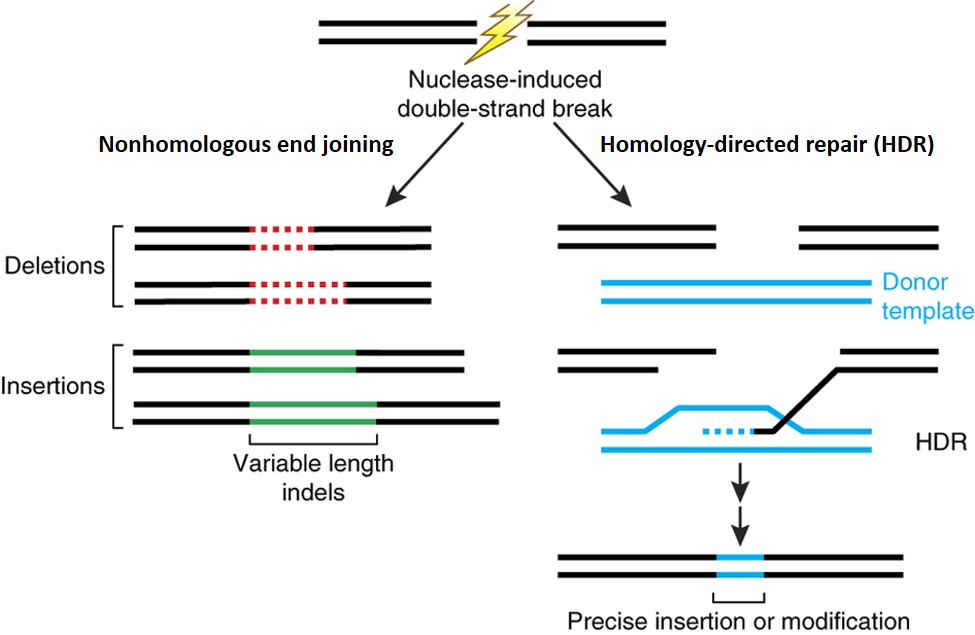
Genome editing offers an approach to introduce targeted edits into the genome. It can be used to introduce targeted knock-outs of specific genes, or perform intraspecies allele substitutions using a donor template with endogenous DNA sequences. Both of these cases do not introduce novel DNA and resemble genetic variations that can be found in the cattle genome. It can also be used to introduce DNA from outside the range of variation found in the cattle genome using a donor template with exogenous DNA sequences, in which case the resulting animal carries exogenous or “transgenic” DNA and is a genetically engineered animal (Figure 1).
Figure 1. Nuclease-induced double-strand breaks (DSBs) can be repaired by nonhomologous end joining (NHEJ) or homology-directed repair (HDR) pathways. Imprecise NHEJ-mediated repair can produce insertion and/or deletion mutations of variable length at the site of the DSB. HDR-mediated repair can introduce precise point mutations or insertions from a single-stranded or double-stranded DNA donor template. Image from Sander and Joung, 2014[1].
Genome editing can be performed in cell culture followed by somatic cell nuclear transfer (SCNT) cloning in which case the genotype of the animal can be known for certain by genotyping or sequencing the cell line prior to cloning. Alternatively, editing can be undertaken in the developing embryo in which case the genotype of the animal will not be known until after it is born[2]. And in that case of an edited embryo the resulting calf may be mosaic – meaning that the cells within the animal have more than one genotype.
In the United States at the current time ALL genome edits in food animals – regardless of the nature of the intended alteration or edit – are regulated as new animal drugs by the FDA. This means that the animal and its products (milk and meat) are considered unapproved animal drugs and are therefore not salable. To obtain a new animal drug approval, or even to obtain permission for the animals to enter the food chain, extensive documentation is required. The following is what was required by the FDA for a “food use authorization” for genome edited cattle under an Investigational New Animal Drug (INAD) and their offspring in 2018:
- Description of animals proposed to enter the food supply:
- Species, class, and number of requested investigational animals to enter the food supply
- Breeding strategy used to produce the offspring
- The purpose of the genetic alteration and the intended function
- Description of the genetic alteration (location in the genome, (impact on protein expression, if any,) intended sequence, etc.) and details of how the genomic alteration was achieved.
- Comparison of the genetically altered sequence to the naturally occurring sequence (sequence alignment)
- Whole Genome Sequencing (WGS) data for:
- the edited parental animal(s) (including information on which animal(s) the data were collected from and the relation of these animal(s) to the offspring)
- the unedited cell line (control)
- the offspring of the genome edited animal
- Health records for the edited cattle and comparison to non-edited cattle
- Provide a nutritional compositional analysis of edible muscle tissues of the investigational animals that will enter food supply including key nutrients. This should be compared with equivalent analysis of a conventional counterpart. Provide information on the methodology used and how the samples were obtained to conduct the meat analysis. Also, information regarding any statistical significance of any observed differences related to natural variation. This analysis may focus on muscle, but if available, can also include data on provide liver, kidney and fat.
- The Codex Guideline (CAC/CL 68-2008) defines key nutrients are those components in a particular food that may have a substantial impact in the overall diet. They may be major constituents (fats, proteins, carbohydrates as nutrients or enzyme inhibitors as anti-nutrients) or minor compounds (minerals, vitamins).
It is worth noting with regard to “off-target” edits or unintended modifications to the genome that a study of whole genome sequence data from 2703 individual cattle in the 1000 Bull Genomes Project revealed more than 86.5 million differences (variants) between different breeds of cattle. These variants included 2.5 million insertions and deletions (indels) of one, or more, base pairs of DNA, and 84 million single nucleotide variants, where one of the four nucleotides making up DNA (A, C, G, T) had been changed to a different one[3]. A small fraction of these mutations have been selected by breeders owing to their beneficial effects on traits of economic importance. None of these naturally-occurring variants are known to produce ill effects on the consumers of milk or beef products. In fact, every meal we have ever consumed is genetically distinct from every other meal in terms of genomic DNA sequences. Genetic variation per se does not pose a unique hazard as it relates to food safety, and it is in fact the fuel that drives animal breeding programs. If an animal that is otherwise healthy and exhibits just the phenotype that is intended by the edit (e.g. changed coat color), then it is hard to envisage a hazard associated with unintended genomic alterations that warrants full genome sequencing. In addition, there is no way to differentiate between naturally occurring genomic alterations and those unintended modifications introduced by editing[4].
Recommendations
According to the International Committee for Animal Recording Section 18 – Guidelines for Breed Associations
- Breed Associations should check the rules of their countries with regard to allowing gene-edited animals in the herd book.
- If an animal has been gene edited it should be recorded against the animal when registered and should appear on the Zootechnical Certificate.
Data Recording
Given this background, edits in the genome of cattle will be referred to as “intended alterations” and it is recommended that the following records should be obtained:
- ID and pedigree of the animal that carries the intended alteration(s)
- Description of the intended alteration (e.g. gene targeted, location in the genome, and intended alteration e.g. allele substitution or SNP change)
- Details of how the intended alteration was achieved (e.g. editing of somatic cells followed by cloning, or introduction of editing regents into zygote and which editing method)
- Timeline of when the intended alteration(s) were made and by which entity (e.g. company or laboratory)
- Data confirming the intended alteration occurred (e.g. Sanger sequencing of targeted genomic region showing knockout or knock-in) showing that the intended alteration(s) that were actually made
- If a donor template plasmid was used in the editing process data should be obtained to confirm the presence or absence of the editing regents in the genome (i.e. data as to whether there was integration of the plasmid backbone into the genome)
- Data documenting whether the animal is heterozygous monollelic edit, a homozygous biallelic edit, a compound heterozygous biallelic edit, or mosaic for the intended alteration (See Figure 2)
- The entity performing the editing should store the information they used to make the animal, and confirm the intended alteration(s) and should provide a certificate to the breeder/breed association confirming the genotype of the animal (e.g. homozygous biallelic polled (PP), heterozygous monoallelic slick (Ss)).
- If an animal is mosaic it would be useful to know if it is germline mosaic as this can affect expected Mendelian inheritance ratios in the offspring. This can take some time as it requires the animal to sexually mature and either test semen or produce offspring, and so perhaps what is important is that breeders know the animal is mosaic so they are aware of this possibility.
Figure 2. Schematic representation of possible outcomes from CRISPR-mediated mutation by cytoplasmic injection of an in vitro fertilized zygote (one cell embryo). 2n = number of homologous chromosomes, i.e. diploid. 2c/4c = number of copies of chromosomes either before or after DNA replication. Image from Supplementary materials of Hennig et al., 2020[5].
References
- ↑ Sander, J.D., Joung, J.K., 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology 32, 347-355. doi:10.1038/nbt.2842.
- ↑ Bishop, T.F., Van Eenennaam, A.L., 2020. Genome editing approaches to augment livestock breeding programs. J Exp Biol 223 doi:10.1242/jeb.207159.
- ↑ Hayes, B.J., Daetwyler, H.D., 2019. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Applications and Outcomes. Annu Rev Anim Biosci 7, 89-102. doi:10.1146/annurev-animal-020518-115024.
- ↑ Van Eenennaam, A.L., Wells, K.D., Murray, J.D., 2019. Proposed U.S. regulation of gene-edited food animals is not fit for purpose. NPJ Sci Food 3, 3. doi:10.1038/s41538-019-0035-y.
- ↑ Hennig, S.L., Owen, J.R., Lin, J.C., Young, A.E., Ross, P.J., Van Eenennaam, A.L., Murray, J.D., 2020. Evaluation of mutation rates, mosaicism and off target mutations when injecting Cas9 mRNA or protein for genome editing of bovine embryos. Scientific Reports 10, 22309. doi:10.1038/s41598-020-78264-8.